Earlier this year, an algorithm was released online — full and completely free — for academics to use. This algorithm represents one of the most important scientific discoveries of our lifetimes, showcasing the growing potential for AI to alter the course of our civilization. Using deep learning and neural networks the algorithm known as Alpha Fold promises to revolutionize the field of biochemistry. It can help us better understand diseases, formulate medicine, and produce solutions to everything from plastic pollution to excess carbon in the atmosphere. By attempting to solve the protein-folding problem we’ve made an exciting and unprecedented breakthrough that will affect the lives of us all.
The protein-folding problem has been an ongoing obstacle for the past 50 years. It first arose in 1972: a new theory proposed that knowing a protein’s amino acid sequence should allow you to fully predict its structure.
Proteins aren’t just fundamental to life, they are responsible for almost all the processes that take place inside a cell. All living organisms rely on these complex molecules. In turn, a protein is made of a chain of 20 different amino acids. The interactions between these amino acids determine how the protein will fold into its 3D shape. The shape of a protein plays a large role in determining its function, hence why in biology there is the saying, “structure is function”. Structure will determine what a protein will do and how it will work. Thus, we might be tempted to conclude that so long as one knows the sequence of a protein’s amino acids, one can determine its final 3D shape. But therein lies the problem.
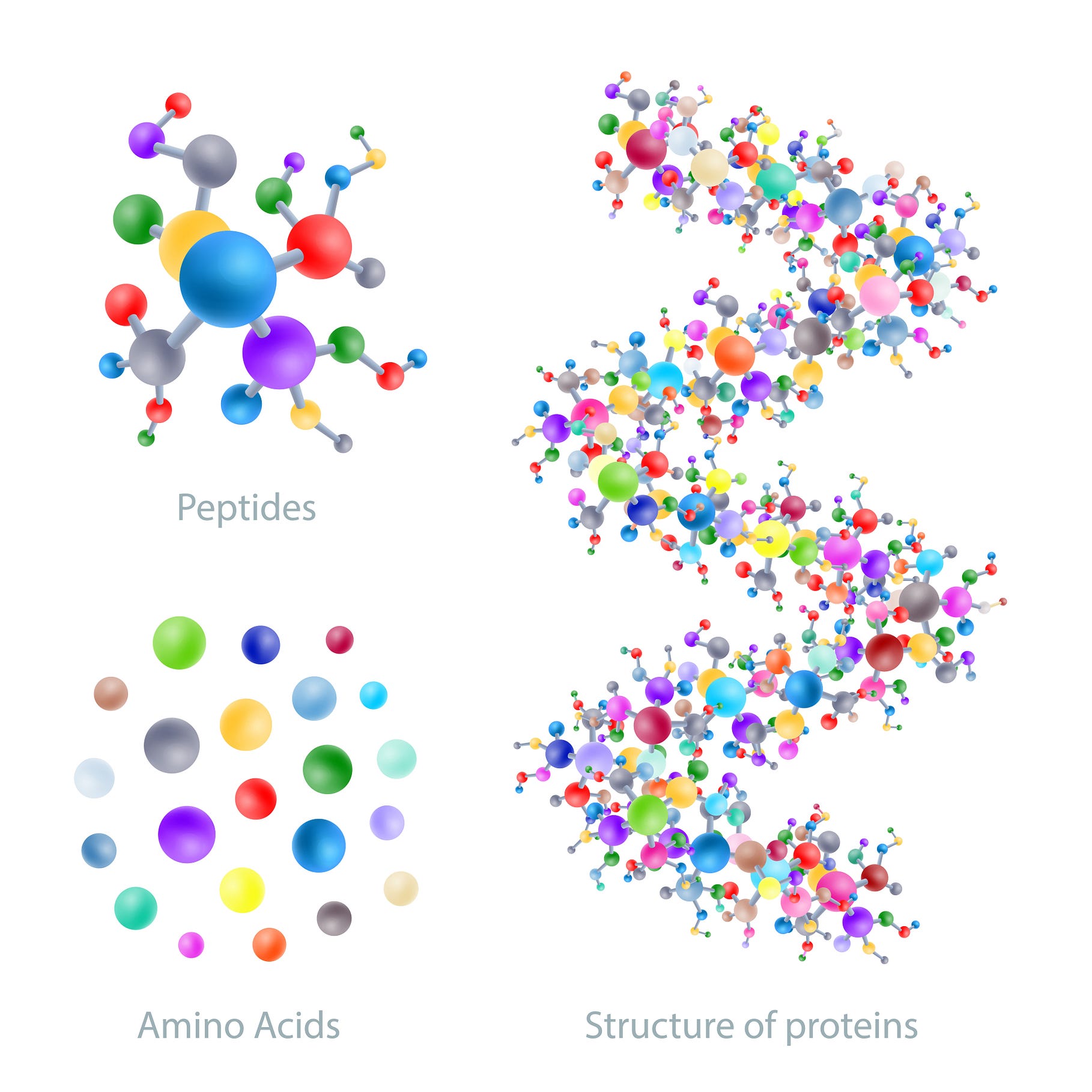
A single protein can be made of up to 2,000 amino acids. Determining all of their possible structures can take longer than the age of the entire universe. This amounts to some 10³⁰⁰ possibilities, meaning that a system capable of predicting how a protein folds will have to use something far more elegant and precise than simple brute force.
Ever since the competition known as the Critical Assessment of protein Structure Prediction (CASP) began in 1994, no competing team has come even close to making accurate predictions. The competition itself consists of hundreds of teams whose algorithms aim to predict the structure of about 100 different proteins from given sequences of amino acids. The structures of the proteins have already been determined experimentally but have not yet been publicly revealed. Predictions made by the teams’ algorithms are then compared to the experimental results and assessed by a panel of judges. Experimental methods used to determine protein structures include X-ray crystallography and cryo-electron microscopy (cryo-EM): well known but expensive methods of research.
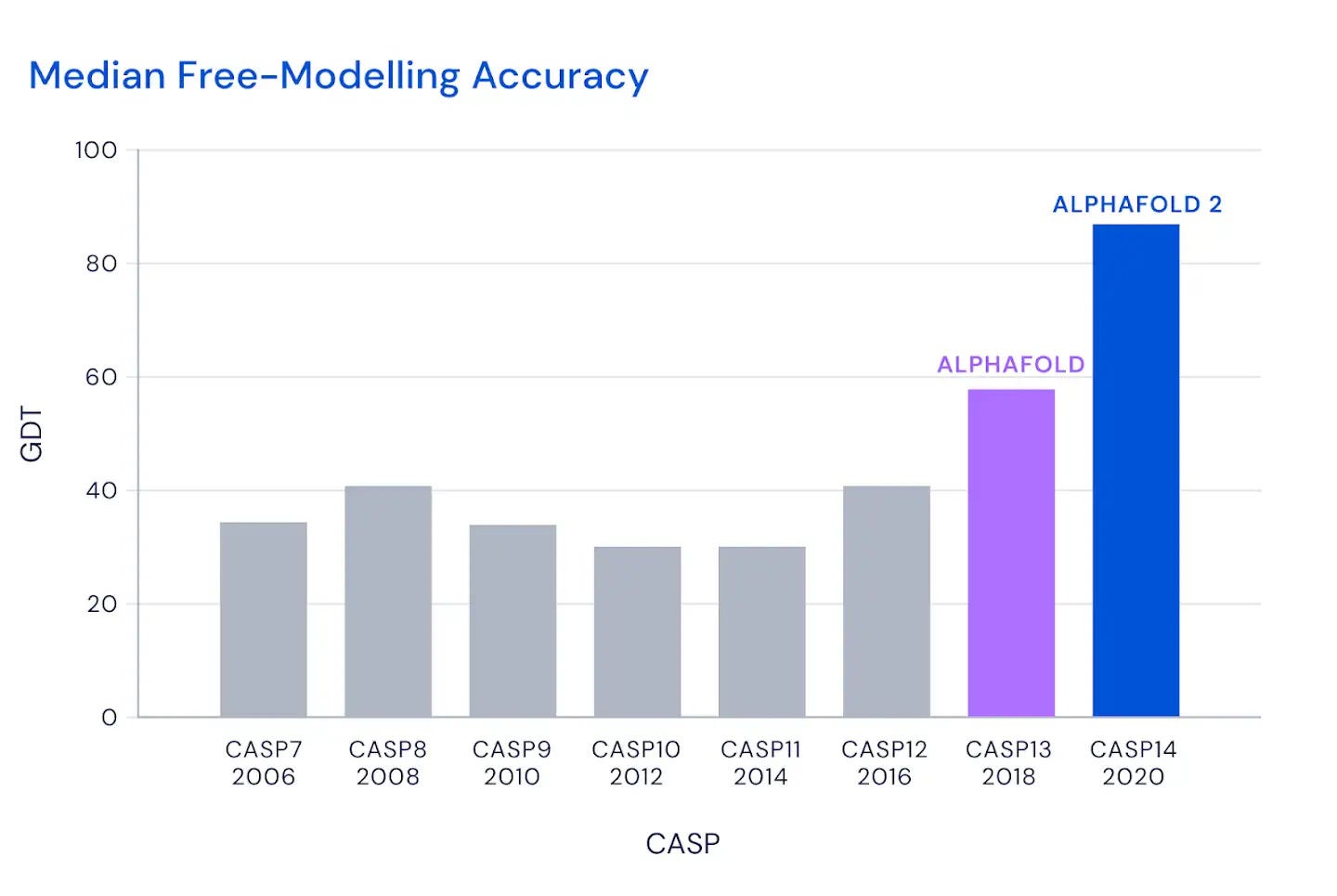
Last year, DeepMind’s Alpha Fold algorithm became the first to make increasingly accurate predictions. The algorithm made strides so astonishing that, to many researchers, the protein-folding problem has essentially been solved.
Alpha Fold’s predictions were on average over 90% accurate in 2020. This is a huge improvement over the average of 40% accuracy achieved by the top performing CASP teams over the last couple of decades. When other teams would score a 75% accuracy rating on moderately difficult proteins, Alpha Fold would score 90%. Even when the algorithm did disagree with experimental results, it wasn’t clear which one was more correct since both allow for a certain margin of error. For many of the Alpha Fold predictions this margin of error was the size of an atom, with the algorithm predicting the exact location of thousands of other atoms in the structure. Overall about two-thirds of the predictions were of the same quality as experimental results.
Figuring out a protein structure can take days or even years using experimental methods like nuclear magnetic resonance, X-ray crystallography, and cryo-EM. They are also more labor-intensive and costly, relying on trial and error and expensive machinery. Yet Alpha Fold isn’t meant to replace them. Instead it’s meant to supplement the work of researchers. Already Alpha Fold has helped scientists find structures of proteins that they’ve been researching for decades, enabling science to move forward where before it had been stalled. The Centre for Enzyme Innovation is using the algorithm to find an enzyme that’ll help us breakdown single use plastics. It’s also inspired other teams — such as one from the University of Washington — to improve upon Alpha Fold in order to make it faster and more energy efficient.
Like the computer programs of the 80’s and 90’s, however, the first iteration of Alpha Fold wasn’t very successful. Its accuracy rating for CASP in 2018 was less than 60%. It wasn’t until the second iteration of Alpha Fold that real progress was made using the help of deep learning. Deep learning is a kind of machine learning that imitates the way a human brain might behave, enabling the machine to learn with far less input from human beings than might be required by traditional machine learning.
Neural networks made of nodes make up the backbone of deep learning. There are at least 3 layers of nodes in a neural network: input, output, and hidden layers in between. Data is shared across nodes in the neural network and the machine then makes predictions which it can check against a dataset. Training data helps the machine improve its predictions. In the case of Alpha Fold’s deep learning network, the training data consisted of folded proteins from the Protein Data Bank. Additionally, instead of only having 1 neural network Alpha Fold has 2 networks that work with one another to fold the protein, render a 3D model, and adjust their alignments of amino acids at the end.

Having made Alpha Fold’s code available for academics to use, the implications could be enormous.
Alpha Fold is working with Drugs for Neglected Diseases initiative (DNDi) and has so far enabled new treatments for illnesses, helping to replace toxic drugs that before could kill 1 in 20 patients with much safer medicine. Alpha Fold has also helped detect and prevent sight-threatening diseases of the eye and is helping in the research of antibiotic resistance. New designs could lead to proteins that breakdown toxic waste, or solutions to the problem of carbon capture. The industries it could revolutionize include medicine, agriculture, bioengineering, biotechnology, and food science, though it’s unclear just how many breakthroughs will actually stem from the release of the Alpha Fold algorithm. It may well take a couple of decades for it to unravel its full potential, changing the world in many unexpected ways.
In the end Alpha Fold’s solution to the protein-folding problem was a case of using machines to understand machines — proteins, after all, are nothing more than microscopic machines programmed to transport oxygen, digest food, and everything in between. It’s a stunning showcase of the power of AI. Artificial Intelligence is this generation’s telescope: an instrument to understand the mysterious phenomena all around us, opening up a new vision of the world.